Searching document text at scale using Azure Cognitive Search
This blog post is accompanied by another post entitled Creating custom skills in Azure Cognitive Search using Azure ML.
This is the second of the two posts. In the first post we explore the creation of a custom skill for use in enriching the document index we’ll be creating here.
Azure cognitive search is a Lucene-based search PaaS service available from Microsoft Azure.
In this post we’ll go through the process of:
- Creating an Azure Storage Account
- Uploading documents to our Azure Storage Account
- Creating an Azure Cognitive Search instance
- Connecting the Azure Cognitive Search instance to our data source
- Connecting the Azure Cognitive Search instance to Cognitive Services
- Defining a skillset including our custom skill
- Indexing our documents
- Querying Azure Search
- Formatting search highlights
The workflow for Azure Cognitive Search looks like the following diagram:
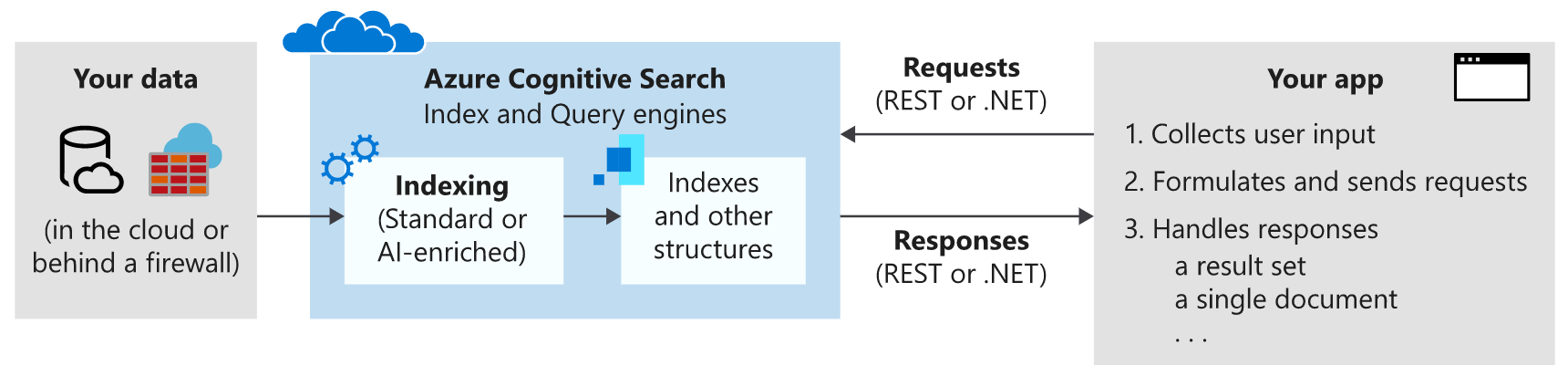
The tools we’ll be using in this post are the Azure CLI and Python to make REST requests to our Cognitive Search instance.
Quick disclaimer: At the time of writing, I am currently a Microsoft Employee
Example Use Case
The example use case to be used here is that we’ll be uploading PDF files, having Azure use the OCR service from Azure Cognitive Services to insert any non-machine readable text, and making the resulting text searchable using Azure Cognitive Search.
The PDF files to be used in this case are a set of 10 PDF files from the year 1980 from the Open Access Journal Nucleic Acids Research (NAR). They can be found here.
Although only 10 PDF files are used here, this can be done at a much larger scale and Azure Cognitive Search supports a range of other file formats including: Microsoft Office (DOCX/DOC, XSLX/XLS, PPTX/PPT, MSG), HTML, XML, ZIP, and plain text files (including JSON).
For reference, the author was previously a Biochemist, with 3 papers published in NAR, though not quite in 1980 – Nucleic Acids Research tends to focus on the field of molecular biology, particularly in relation to DNA and RNA, and proteins and other molecules that interact with these nucleic acids.
An example of what one of our files looks like can be seen here:

Creating an Azure Storage Account
We’ll start by creating an Azure Storage Account in which to store our documents.
Using the Azure CLI, create a resource group (if not already created), I’ve named mine azure-search-nar-demo:
az group create --name azure-search-nar-demo --location westeurope
Then, again using the Azure CLI, create a storage account, this one is named narsearchdemostorage:
az storage account create --name narsearchdemostorage --resource-group azure-search-nar-demo --location westeurope
We’ll need the connection string for this account, to retrieve it for this account run the following:
az storage account show-connection-string --name narsearchdemostorage
Uploading PDF files to Azure Storage Account
We’ll be using the following two external python packages in this post, so I’d recommend pip installing them into a python virtual environment:
pip install azure-storage-blob
pip install requests
Let’s go ahead and import the python modules we’ll need for this section:
import os
from azure.storage.blob import BlobServiceClient, BlobClient, ContainerClient
Now we’ll instantiate the blob service client using the connection string we got from the Azure CLI in the previous section:
connection_string = "<azure_storage_connection_string>"
blob_service_client = BlobServiceClient.from_connection_string(connection_string)
Within this storage account, we’ll need to create a BLOB container, this is done as follows:
container_name = "nar-pdf-container"
container_client = blob_service_client.create_container(container_name)
Before we upload our files, let’s list the directory and take a look at what files we’ll be uploading:
os.listdir('./pdfs')
We can now iterate through these files and upload each of them to our storage account (it may be easier for larger numbers of files to use the AzCopy tool or Azure Storage Explorer).
for pdf in os.listdir('./pdfs'):
blob_client = blob_service_client.get_blob_client(
container=container_name,
blob=pdf
)
with open(os.path.join('.', 'pdfs', pdf), "rb") as data:
blob_client.upload_blob(data)
To check our files have uploaded, we can list the files in the container:
blob_list = container_client.list_blobs()
for blob in blob_list:
print(blob.name)
Create Azure Cognitive Search Service
We’ll now create our Azure Cognitive Search service, I’ve called mine nar-demo-search – Using Azure CLI run:
az search service create --name nar-demo-search --resource-group azure-search-nar-demo --location westeurope --sku standard
SKU options can be found here.
This can take a while so go and grab a coffee while this deploys.
Once deployed, we’ll need the admin key for this Azure Cognitive Search service instance, to access this you can run using Azure CLI:
az search admin-key show --service-name nar-demo-search --resource-group azure-search-nar-demo
Connect Azure Cognitive Search Service to BLOB storage
In the cell below, we’ll import modules and define variables that we’ll need for each of the Azure Cognitive Search Service REST requests for the rest of this blog post.
For each request to Azure Cognitive Search, you must provide an API Version, in this post, we’re using the latest stable version, which is "2019-05-06".
import requests
import json
service_name = "nar-demo-search"
api_version = "2019-05-06"
headers = {
'Content-Type': 'application/json',
'api-key': "<azure_cognitive_search_admin_key>"
}
Now we can connect to our data source, we’ll need to provide the Azure Storage Account connection string we previously used to connect to our Storage account and we’ll need to give this data source a name.
We also provide the container in which our data is stored.
datasource_name = "blob-datasource"
uri = f"https://{service_name}.search.windows.net/datasources?api-version={api_version}"
body = {
"name": datasource_name,
"type": "azureblob",
"credentials": {"connectionString": connection_string},
"container": {"name": container_name}
}
resp = requests.post(uri, headers=headers, data=json.dumps(body))
print(resp.status_code)
print(resp.ok)
Connect Azure Cognitive Search Service to Cognitive Services
Next we’re going to define the AI skillset which will be used to enrich our search index.
But first, in order to do this, it’s advisable to create an Azure Cognitive Services instance, otherwise your AI enrichment capabilities will be severely limited in scope. This Azure Cognitive Services instance must be in the same region as your Azure Cognitive Search instance, and can be created from the CLI, I’ve named mine nar-demo-cognitive-services:
az cognitiveservices account create --name nar-demo-cognitive-services --kind CognitiveServices --sku S0 --resource-group azure-search-nar-demo --location westeurope
We’ll need the account key for this Azure Cognitive Services instance in order to connect our Azure Cognitive Search instance to it, we can retrieve the key using the Azure CLI:
az cognitiveservices account keys list --name nar-demo-cognitive-services --resource-group azure-search-nar-demo
Create Azure Cognitive Search Skillset
To enrich our Azure Cognitive Search Index with an AI skillset, we’ll need to define a skillset.
We’ll be using a combination of skills that utilise Azure Cognitive Services and our own custom skill deployed in Azure Machine Learning. The skills we’ll be using are:
- OCR to extract text from image
- Merge text extracted from OCR into the correct place in documents
- Detect language
- Split text into pages if not already done so
- Key phrase extraction (has a maximum character limit so requires text to be split into pages)
- Our Custom Skill to extract genetic codes.
The skillset definition is a JSON object and each of the skills defined take one or more fields as input and provide one or more fields as output.
We’ll need to provide the Cognitive Search API with the key to our Cognitive Services account in order to use it, so make sure you put this in your skillset definition.
We’ll also need the URI and key for our Azure Cognitive Search Skill API that we previously deployed in Azure Machine Learning, so make sure to update the skillset definition below too.
skillset = {
"description": "Extract text from images and merge with content text to produce merged_text. Also extract key phrases from pages",
"skills":
[
{
"description": "Extract text (plain and structured) from image.",
"@odata.type": "#Microsoft.Skills.Vision.OcrSkill",
"context": "/document/normalized_images/*",
"defaultLanguageCode": "en",
"detectOrientation": True,
"inputs": [
{
"name": "image",
"source": "/document/normalized_images/*"
}
],
"outputs": [
{
"name": "text"
}
]
},
{
"@odata.type": "#Microsoft.Skills.Text.MergeSkill",
"description": "Create merged_text, which includes all the textual representation of each image inserted at the right location in the content field.",
"context": "/document",
"insertPreTag": " ",
"insertPostTag": " ",
"inputs": [
{
"name":"text", "source": "/document/content"
},
{
"name": "itemsToInsert", "source": "/document/normalized_images/*/text"
},
{
"name":"offsets", "source": "/document/normalized_images/*/contentOffset"
}
],
"outputs": [
{
"name": "mergedText", "targetName" : "merged_text"
}
]
},
{
"@odata.type": "#Microsoft.Skills.Text.LanguageDetectionSkill",
"inputs": [
{ "name": "text", "source": "/document/content" }
],
"outputs": [
{ "name": "languageCode", "targetName": "languageCode" }
]
},
{
"@odata.type": "#Microsoft.Skills.Text.SplitSkill",
"textSplitMode" : "pages",
"maximumPageLength": 4000,
"inputs": [
{ "name": "text", "source": "/document/content" },
{ "name": "languageCode", "source": "/document/languageCode" }
],
"outputs": [
{ "name": "textItems", "targetName": "pages" }
]
},
{
"@odata.type": "#Microsoft.Skills.Text.KeyPhraseExtractionSkill",
"context": "/document/pages/*",
"inputs": [
{ "name": "text", "source": "/document/pages/*" },
{ "name":"languageCode", "source": "/document/languageCode" }
],
"outputs": [
{ "name": "keyPhrases", "targetName": "keyPhrases" }
]
},
{
"@odata.type": "#Microsoft.Skills.Custom.WebApiSkill",
"description": "This skill extracts genetic codes from text",
"uri": "<amls_skill_uri>",
"context": "/document/pages/*",
"httpHeaders": {
"Authorization": "Bearer <amls_skill_key>"
},
"inputs": [
{ "name" : "text", "source": "/document/pages/*"}
],
"outputs": [
{ "name": "genetic_codes", "targetName": "genetic_codes" }
]
}
],
"cognitiveServices": {
"@odata.type": "#Microsoft.Azure.Search.CognitiveServicesByKey",
"description": "NAR Demo Cognitive Services",
"key": "<azure_cognitive_services_key>"
}
}
The skillset is created through a PUT request to our Azure Cognitive Search skillsets REST endpoint.
The data above is serialised to a JSON string before being provided as the body of the request:
skillset_name = 'nar-demo-skillset'
uri = f"https://{service_name}.search.windows.net/skillsets/{skillset_name}?api-version={api_version}"
resp = requests.put(uri, headers=headers, data=json.dumps(skillset))
print(resp.status_code)
print(resp.ok)
Create Azure Cognitive Search Index
The index is the definition of fields that will be returned, as well as metadata such as the data type of this field, whether it is a key or not, and whether it is searchable, filterable, facetable or sortable.
We’ll return the id, metadata_storage_name, content, languageCode, keyPhrases, genetic_codes, and merged_text field and make all of them searchable.
index = {
"fields": [
{
"name": "id",
"type": "Edm.String",
"key": True,
"searchable": False,
"filterable": False,
"facetable": False,
"sortable": True
},
{
"name": "metadata_storage_name",
"type": "Edm.String",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": True
},
{
"name": "content",
"type": "Edm.String",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": False
},
{
"name": "languageCode",
"type": "Edm.String",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": False
},
{
"name": "keyPhrases",
"type": "Collection(Edm.String)",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": False
},
{
"name": "genetic_codes",
"type": "Collection(Edm.String)",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": False
},
{
"name": "merged_text",
"type": "Edm.String",
"searchable": True,
"filterable": False,
"facetable": False,
"sortable": False
}
]
}
Just as with our skillset, the index is created through a PUT request to our Azure Cognitive Search indexes REST endpoint.
The data above is serialised to a JSON string before being provided as the body of the request:
index_name = 'nar-demo-index'
uri = f"https://{service_name}.search.windows.net/indexes/{index_name}?api-version={api_version}"
resp = requests.put(uri, headers=headers, data=json.dumps(index))
print(resp.status_code)
print(resp.ok)
Create Azure Cognitive Search Indexer
We’ll now create our indexer, which runs through our data and indexes it as we’ve defined above using the data source, index and skillset given.
We can set maxFailedItems to maxFailedItemsPerBatch to -1 if we want the indexer to continue through until reaching the end of all the documents regardless of how many failures it encounters.
The storage path of the file within the storage container will be base 64 encoded and act as a key as it will be unique for each document, regardless of whether two files have the same filename.
indexer_name = "nar-demo-indexer"
indexer = {
"name": indexer_name,
"dataSourceName" : datasource_name,
"targetIndexName" : index_name,
"skillsetName" : skillset_name,
"fieldMappings" : [
{
"sourceFieldName" : "metadata_storage_path",
"targetFieldName" : "id",
"mappingFunction" : {"name": "base64Encode"}
},
{
"sourceFieldName" : "metadata_storage_name",
"targetFieldName" : "metadata_storage_name",
},
{
"sourceFieldName" : "content",
"targetFieldName" : "content"
}
],
"outputFieldMappings" :
[
{
"sourceFieldName" : "/document/merged_text",
"targetFieldName" : "merged_text"
},
{
"sourceFieldName" : "/document/pages/*/keyPhrases/*",
"targetFieldName" : "keyPhrases"
},
{
"sourceFieldName" : "/document/pages/*/genetic_codes/*",
"targetFieldName" : "genetic_codes"
},
{
"sourceFieldName": "/document/languageCode",
"targetFieldName": "languageCode"
}
],
"parameters":
{
"maxFailedItems": 1,
"maxFailedItemsPerBatch": 1,
"configuration":
{
"dataToExtract": "contentAndMetadata",
"parsingMode": "default",
"firstLineContainsHeaders": False,
"delimitedTextDelimiter": ",",
"imageAction": "generateNormalizedImages"
}
}
}
You may be sensing a theme now to creating associated resources for our Azure Cognitive Search instance. Just as with our skillset and index, the indexer is created through a PUT request to our Azure Cognitive Search indexers REST endpoint.
The data above is serialised to a JSON string before being provided as the body of the request. This will kick off the indexing on our PDF files.
We have not defined a schedule in our indexer so it will just run once but if you wish to provide a schedule that will check for updates to the index you can, for example, every 2 hours would be:
"schedule" : { "interval" : "PT2H" }
And 5 minutes, the shortest interval, would be:
"schedule" : { "interval" : "PT5M" }
indexer_name = 'nar-demo-indexer'
uri = f"https://{service_name}.search.windows.net/indexers/{indexer_name}?api-version={api_version}"
resp = requests.put(uri, headers=headers, data=json.dumps(indexer))
print(resp.status_code)
print(resp.ok)
To check on the status of our indexer, you can run the following cell.
The JSON response has more information for if you want to know how many files the indexer has completed and how many is left to go.
uri = f"https://{service_name}.search.windows.net/indexers/{indexer_name}/status?api-version={api_version}"
resp = requests.get(uri, headers=headers)
print(resp.status_code)
print(resp.json().get('lastResult').get('status'))
print(resp.ok)
Querying the Azure Cognitive Search Index
Now that our documents have been indexed, we can start to query our Azure Cognitive Search index.
I mentioned that the journal NAR is focused on the research of nucleic acids DNA and RNA, so we should expect that if we were to search for, say, the term “RNA” we should get back most, if not all, of our documents.
# Base URL
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
# API version is required
url += '?api-version={}'.format(api_version)
# Search query of "RNA"
url += '&search=RNA'
# Return the count of the results
url += '&$count=true'
print(url)
resp = requests.get(url, headers=headers)
print(resp.status_code)
The 200 status code is a good start. To get the search results from our response, we must call the json() method of our response object to de-serialise the JSON string object that is returned in the body of our response.
search_results = resp.json()
search_results.keys()
This object is a dict with 3 keys:
@odata.context– The index and documents that were searched@odata.count– The count of documents returned from our index search queryvalue– The output fields of the index search query
search_results['@odata.context']
search_results['@odata.count']
We can see in the cells above and below that we are, indeed, returned all 10 documents by querying our index with the word “RNA”.
len(search_results['value'])
For each item in value, we are returned the fields that we defined in our index, as well as a search score:
search_results['value'][0].keys()
We can take a look at the results of our search by looking at the PDF name and search score.
By default, the output of the query is ordered by search score and we have not defined any weightings to fields but this is something that can also be defined:
for result in search_results['value']:
print('PDF Name: {}, Search Score {}'.format(result['metadata_storage_name'], result['@search.score']))
In the cell below, we take a look at the beginning of the merged text of the highest scoring search result. Given that the name of this article is “Evidence for the role of double-helical structures in the maturation of Sinian Virus-40 messenger RNA”, it’s unsurprising that this is the highest scoring search result when searching for “RNA”.
print(search_results['value'][0]['merged_text'][:350])
Let’s now take a look at a query that won’t just return every document that we uploaded.
Transcription is the mechanism by which DNA is encoded into RNA as the initial step in gene expression, ready to be translated into proteins.
Let’s do a search for “transcription” and see how many results our query returns.
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=transcription'
url += '&$count=true'
print(url)
resp = requests.get(url, headers=headers)
print(resp.status_code)
In the following cell, we find that the term transcription is found in 4 out of our 10 documents.
search_results = resp.json()
search_results['@odata.count']
for result in search_results['value']:
print('PDF Name: {}, Search Score {}'.format(result['metadata_storage_name'], result['@search.score']))
When looking at our highest result, we can be buoyed once more by the fact that the highest scoring search result is likely to be highly related to transcription – this journal article is about the regions of the ovalbumin gene that are responsible for controlling gene expression and is led by noted Molecular Geneticist Pierre Chambon, whose work has focused on gene transcription.
print(search_results['value'][0]['merged_text'][:715])
Custom Skill Output
In the other blog post Creating custom skills in Azure Cognitive Search using Azure ML Service we defined a custom skill to extract genetic codes from journal articles when the index is created.
The genetic codes are returned in our search results.
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=*'
url += '&$count=true'
resp = requests.get(url, headers=headers)
search_results = resp.json()
for result in search_results['value']:
print(result['metadata_storage_name'])
print(result['genetic_codes'])
We can provide a search query to search within collections using e.g. search=collection_name/any(x: x eq 'foo') to search for foo within the collection collection_name.
So if we search for one of the genetic codes above, this journal article will be top of our search results.
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += "&search=genetic_codes/any(c: c eq 'TAGGAT')"
url += '&$count=true'
resp = requests.get(url, headers=headers)
search_results = resp.json()
search_results['value'][0]['genetic_codes']
Pagination of Search Results
As we might not want all results all of the time from our search, and indeed by default only the first 50 results will be returned, we might want to paginate our results, to allow users to explore results in batches.
Let’s say we want to search for RNA, which we already know will find 10 results, but we only want to display 5 results on a page. We can do this by supplying to our URL a $top parameter.
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=RNA'
url += '&$count=true'
url += '&$top=5'
print(url)
resp = requests.get(url, headers=headers)
print(resp.status_code)
search_results = resp.json()
print("Results Found: {}, Results Returned: {}".format(search_results['@odata.count'], len(search_results['value'])))
print("Highest Search Score: {}".format(search_results['value'][0]['@search.score']))
That’s perfect for our first page, but what about the second page? We need a way of skipping the first 5 results and showing the next 5 highest scoring results.
We can do this by supplying both $top and $skip parameters to our URL.
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=RNA'
url += '&$count=true'
url += '&$top=5'
url += '&$skip=5'
print(url)
resp = requests.get(url, headers=headers)
print(resp.status_code)
search_results = resp.json()
print("Results Found: {}, Results Returned: {}".format(search_results['@odata.count'], len(search_results['value'])))
print("Highest Search Score: {}".format(search_results['value'][0]['@search.score']))
Article Highlights from Search Results
One of my favourite features of Azure Cognitive Search is the ability to highlight parts of the document that are relevant to our search results. Combined with the above, this really helps turn our API into a proper search engine.
We can request highlights on a particular field by adding the highlight parameter to our URL as below. Let’s highlight the results we had from our search for “transcription”
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=transcription'
url += '&$count=true'
url += '&highlight=merged_text'
resp = requests.get(url, headers=headers)
print(resp.status_code)
search_results = resp.json()
This will extract relevant parts of the journal article regarding “transcription”.
We can see in the cell below, which shows the highlights from the search result, that this will return a list of highlights from the article and even provides HTML em tags for the word transcription.
search_results['value'][0]['@search.highlights']['merged_text']
If we’re using a jupyter notebook, we can display these highlights with the word transcription emphasised through italicisation.
from IPython.display import display, HTML
for highlight in search_results['value'][0]['@search.highlights']['merged_text']:
display(HTML(highlight))
But that’s not all, if we wanted to supply our own tags to, for example, highlight instead of italicise these highlights we can supply our own HTML tags to surround the query phrase.
In this case we’re providing span tags with some inline CSS.
import urllib
url = 'https://{}.search.windows.net/indexes/{}/docs'.format(service_name, index_name)
url += '?api-version={}'.format(api_version)
url += '&search=transcription'
url += '&$count=true'
url += '&highlight=merged_text'
url += '&highlightPreTag=' + urllib.parse.quote('<span style="background-color: #f5e8a3">', safe='')
url += '&highlightPostTag=' + urllib.parse.quote('</span>', safe='')
resp = requests.get(url, headers=headers)
print(url)
print(resp.status_code)
search_results = resp.json()
Now when we display our search results, the output looks a little bit more like that of a regular search engine.
Feel free to explore your Azure Cognitive Search endpoints and don’t forget to delete your resource group when you’re finished exploring.
for result in search_results['value']:
display(HTML('<h4>' + result['metadata_storage_name'] + '</h4>'))
for highlight in result['@search.highlights']['merged_text']:
display(HTML(highlight))