Decision Tree Classifier in Python using Scikit-learn
Decision Trees can be used as classifier or regression models.
A tree structure is constructed that breaks the dataset down into smaller subsets eventually resulting in a prediction. There are decision nodes that partition the data and leaf nodes that give the prediction that can be followed by traversing simple IF..AND..AND….THEN logic down the nodes.
The root node (the first decision node) partitions the data based on the most influential feature partitioning. There are 2 measures for this, Gini Impurity and Entropy.
Entropy
The root node (the first decision node) partitions the data using the feature that provides the most information gain.
Information gain tells us how important a given attribute of the feature vectors is.
It is calculated as:
$\text{Information Gain} = \text{entropy(parent)} – \text{[average entropy(children)]} $
Where entropy is a common measure of target class impurity, given as:
$ Entropy = \Sigma_i – p_i \log_2 p_i $
where i is each of the target classes.
Gini Impurity
Gini Impurity is another measure of impurity and is calculated as follows:
$ Gini = 1 – \Sigma_i p_i^2 $
Gini impurity is computationally faster as it doesn’t require calculating logarithmic functions, though in reality which of the two methods is used rarely makes too much of a difference.
Predicting Survival in the Titanic Data Set
We’ll be using a decision tree to make predictions about the Titanic data set from Kaggle. This data set provides information on the Titanic passengers and can be used to predict whether a passenger survived or not.
import pandas as pd
df = pd.read_csv('data/titanic.csv', index_col='PassengerId')
print(df.head())
We will be using Pclass, Sex, Age, SibSp (Siblings aboard), Parch (Parents/children aboard), and Fare to predict whether a passenger survived.
df = df[['Pclass', 'Sex', 'Age', 'SibSp', 'Parch', 'Fare', 'Survived']]
We need to convert ‘Sex’ into an integer value of 0 or 1.
df['Sex'] = df['Sex'].map({'male': 0, 'female': 1})
We will also drop any rows with missing values.
df = df.dropna()
X = df.drop('Survived', axis=1)
y = df['Survived']
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
from sklearn import tree
model = tree.DecisionTreeClassifier()
Let’s take a look at our model’s attributes
model
Defining some of the attributes like max_depth, max_leaf_nodes, min_impurity_split, and min_samples_leaf can help prevent overfitting the model to the training data.
First we fit our model using our training data.
model.fit(X_train, y_train)
Then we score the predicted output from model on our test data against our ground truth test data.
y_predict = model.predict(X_test)
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_predict)
We see an accuracy score of ~83.2%, which is significantly better than 50/50 guessing.
Let’s also take a look at our confusion matrix:
from sklearn.metrics import confusion_matrix
pd.DataFrame(
confusion_matrix(y_test, y_predict),
columns=['Predicted Not Survival', 'Predicted Survival'],
index=['True Not Survival', 'True Survival']
)
If we have graphviz installed http://www.graphviz.org/, we can export our decision tree so we can explore the decision and leaf nodes.
tree.export_graphviz(model.tree_, out_file='tree.dot', feature_names=X.columns)
We can then convert this dot file to a png file.
from subprocess import call
call(['dot', '-T', 'png', 'tree.dot', '-o', 'tree.png'])
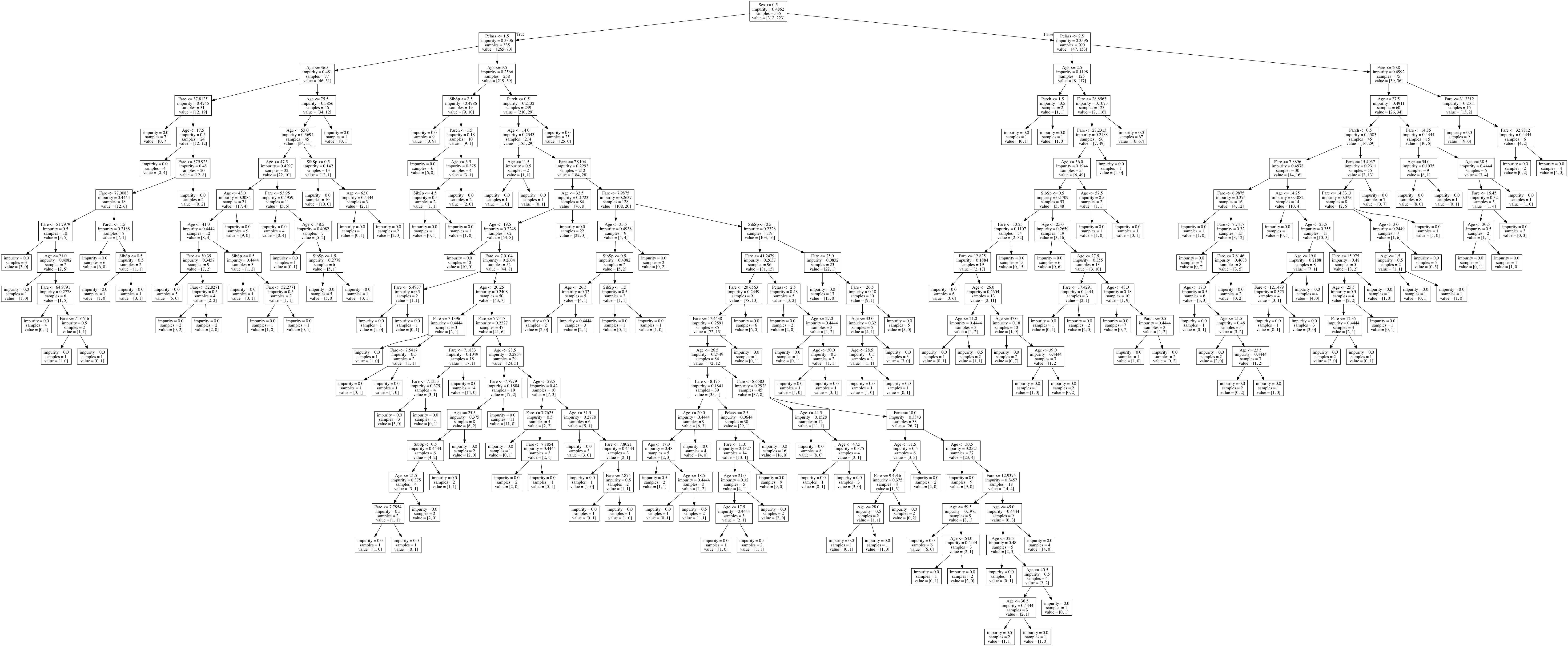
We can then view our tree, which looks like this (Click to view full):
The root node, with the most information gain, tells us that the biggest factor in determining survival is Sex.
If we zoom in on some of the leaf nodes, we can follow some of the decisions down.
We have already zoomed into the part of the decision tree that describes males, with a ticket lower than first class, that are under the age of 10.
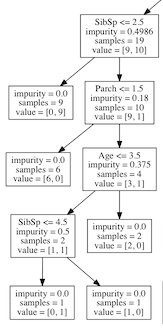
The impurity is the measure as given at the top by Gini, the samples are the number of observations remaining to classify and the value is the how many samples are in class 0 (Did not survive) and how many samples are in class 1 (Survived).
Let’s follow this part of the tree down, the nodes to the left are True and the nodes to the right are False:
- We see that we have 19 observations left to classify: 9 did not survive and 10 did.
- From this point the most information gain is how many siblings (SibSp) were aboard.
A. 9 out of the 10 samples with less than 2.5 siblings survived.
B. This leaves 10 observations left, 9 did not survive and 1 did. - 6 of these children that only had one parent (Parch) aboard did not survive.
- None of the children aged > 3.5 survived
- Of the 2 remaining children, the one with > 4.5 siblings did not survive.